Ozgene big genomic replacement (OzBIG)
Ozgene big genomic replacement (OzBIG)
Mouse humanizations up to 240 kb
Large genomic humanizations up to 240 kb by the replacement of a murine gene/s with its human orthologue. This technology is called ‘Ozgene Big Genomic Replacement’ – or OzBIG for short.
Reduced timelines. Reduced project costs.

Conventional gene targeting
- Targeting vector payload of 15 kb max
- 5 serial gene targeting events
- Time to humanized F1 mice > 3 years
- Cost > 5 projects
- High risk – due to ES cell culture time
OzBIG gene targeting
- Targeting vector payload of 240 kb max
- 1 targeting event
- Time to humanized F1 mice ≈ 7 months
- Cost 1 project
- Risk – low
Conventional gene targeting
- Targeting vector payload of 15 kb max
- 5 serial gene targeting events
- Time to humanized F1 mice > 3 years
- Cost > 5 projects
- High risk – due to ES cell culture time
OzBIG gene targeting
- Targeting vector payload of 240 kb max
- 1 targeting event
- Time to humanized F1 mice ≈ 7 months
- Cost 1 project
- Risk – low
Large complex transgene structures
Large genomic replacements allow for the complete humanization of most genes and the expression of the human gene from its accompanying cis-regulatory elements. They have multiple valuable applications in the medical research field. OzBIG can also be used to build large non-humanized, transgenics like complex reporters and synthetic expression systems.
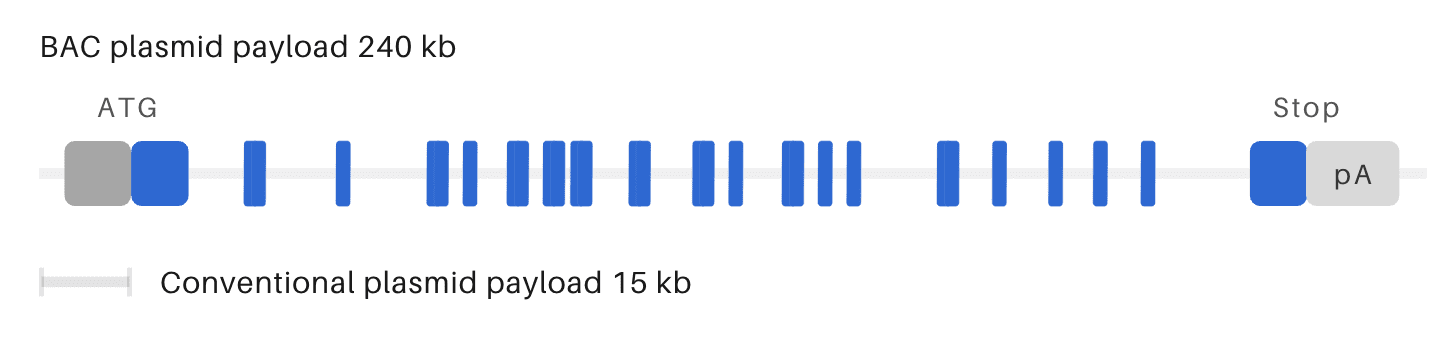
OzBIG humanized mouse
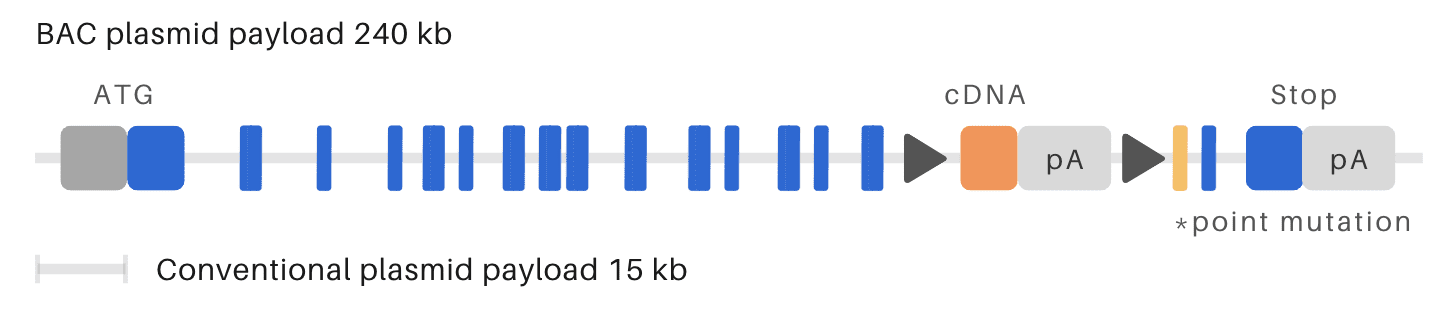
OzBIG conditional point mutation KI
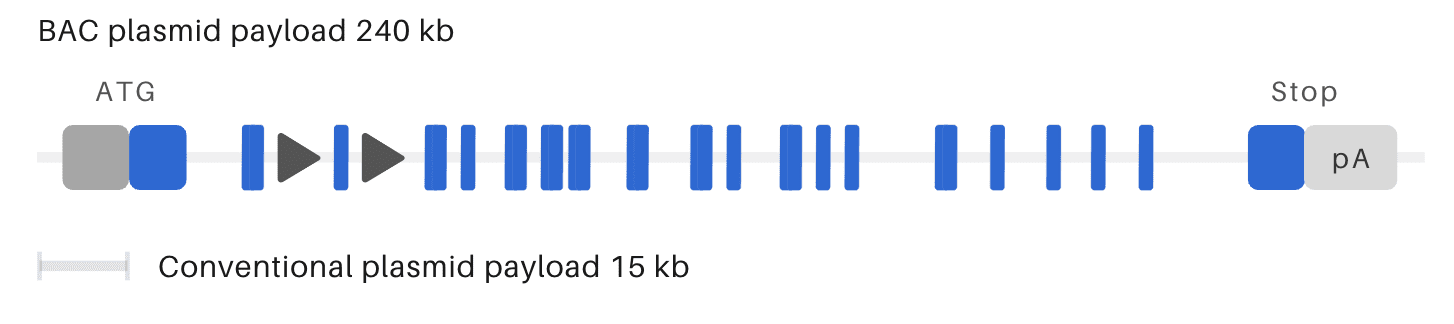
OzBIG humanized conditional KO
Expanded humanization gene set accessibility
Humanisation by genomic replacement
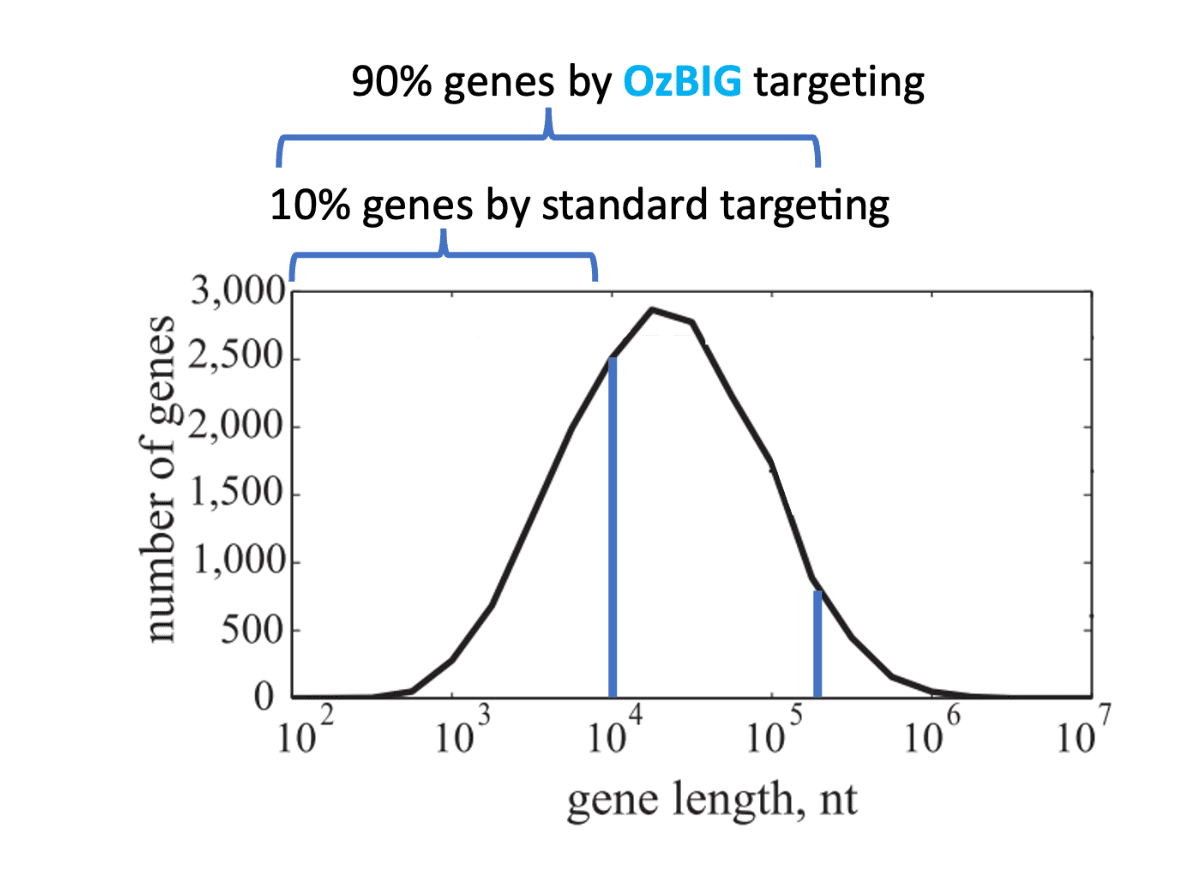
OzBIG vastly expands the gene set for humanisation of a mouse by enabling the insertion of 90% of genes in the human genome, compared to 10% of human genes accessible through standard gene targeting. OzBIG combines BAC gene targeting technology with a screening platform developed at Ozgene to efficiently generate ES cell clones carrying modifications with a current upper limit of 240 kb. Coupled with Ozgene’s goGermline™ embryos, which ensure germline transmission in the first litter, large humanization mouse models are generated in the fastest possible timeframe.
Applications for humanizations
Mice carrying large genomic humanizations can be useful as models of human genetic disease as well as in the development of corrective gene therapies. Models containing human genomic sequences with clinically relevant mutations can be used as experimental targets for the application of Cas9-based prime editing technologies to correct genomic mutations for the treatment of human genetic disease.
Genomic humanizations also permit the expression of a full-length humanized transcript that can be useful for the study of therapeutic agents aimed at treating genetic human diseases by correcting defects in transcript splicing, modulating transcript levels or by altering the transcription or regulation of RNA.
These large genetic modifications also allow for humanizing multiple linked genes and/or large modifications, such as the addition of reporters or conditional elements.
Mice expressing human proteins can also be useful in the study of therapeutic drugs or antibodies that are tailored to interact specifically with the target human protein of interest.

Read More
Get in touch
Take your humanized models to the next level. Reach out to us for a confidential discussion about your mouse model needs. Request a free quote today.
Please fill out the form and we will respond to your query within two business days. Alternatively, visit our contact page for more ways that you can get in touch with us.
‘I selected Ozgene because of the reputation, scientific quality and compatible prices. The best aspects of Ozgene service were professionalism, friendly staff and regular updates on work progress.’
– Prof. Dee Harrison-Findik, University of Nebraska, USA

